Build Tutorial¶
In a nutshell¶
The BUILD scripts configure cMonkey2 to run on an SGE managed cluster. In the process, they change several cMonkey2 parameters to generate variability in each run. They also choose experiments to include in each run according to user-provided annotations.
Requirements¶
User must supply several files, including:
ratios: a Tab-delimited or Comma-separated file containing a matrix of gene expression values across all conditions.blocks: a Comma-separated file containing annotations for each condition.inclusion_blocks: a Comma-separated file containing groups of blocks to be co-included in runs.exclusion_blocks: a Comma-separated file containing groups of blocks to be co-excluded in runs.
Optionally:
pipeline: a JSON file containing custom scoring pipeline. Currently only set-enrichment pipeline is supportedsetenrich_files: Comma-separated files containing set-enrichment sets. Multiple files should be separated by a comma.
The format for each of these files will be described in detail below.
We have provided example files for building an Mycobacterium Tuberculosis ensemble. The data come from the following publications:
Additionally, the Python modules described in this documentation’s dependencies are required.
Scripts¶
egrin2-make_ensemble: The control function for BUILD scripts. Writes QSub script.cMonkeyIniGen.py: Templating function to generate cMonkey2 initialization (.ini) files.ensemblePicker.py: Picks experiments to include in a run given user-supplied experimental blocks
BUILD an EGRIN 2.0 ensemble¶
In this tutorial we will BUILD an ensemble for Mycobacterium tuberculosis using several example files, which we provide here
STEP 1: Generate required input files¶
First, let’s explore the required files.
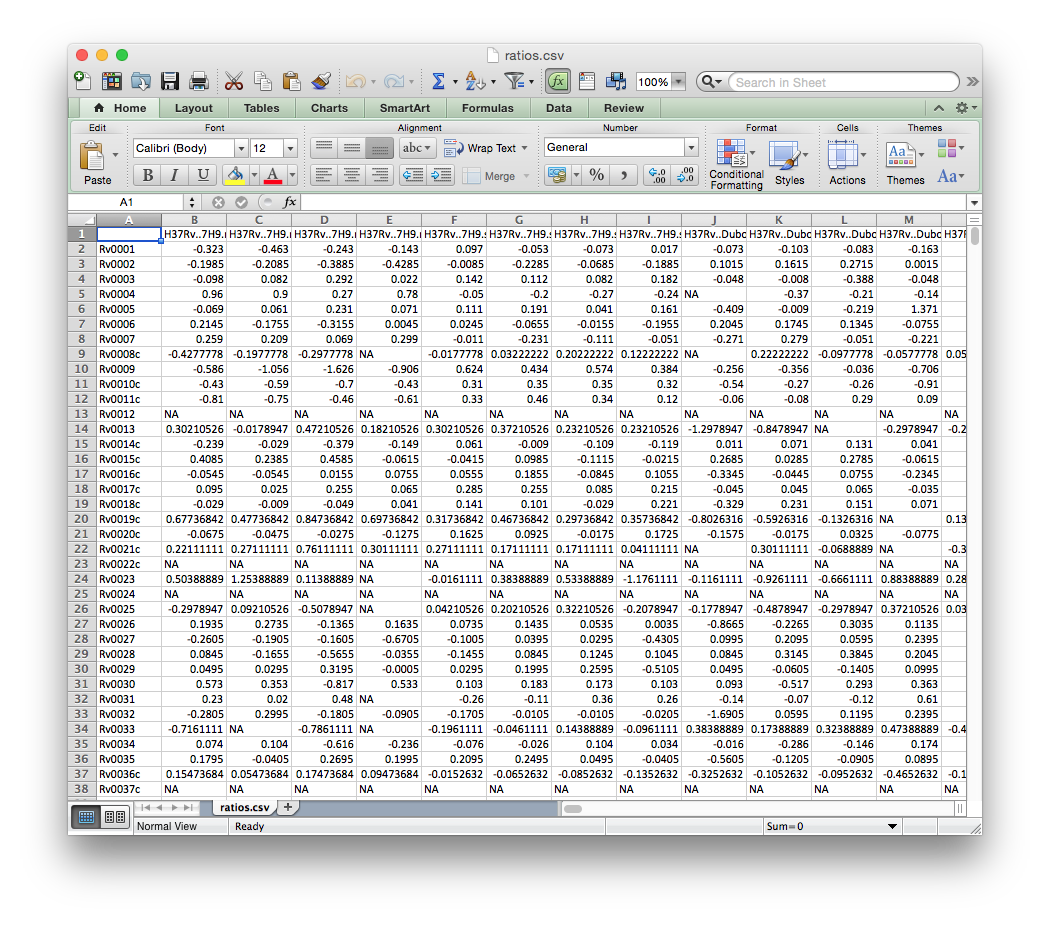
ratios.csv¶
A Tab-delimited or Comma-separated file containing a matrix of gene expression values across all conditions. Rows should correspond to genes and columns to individual conditions, as in the example file below.
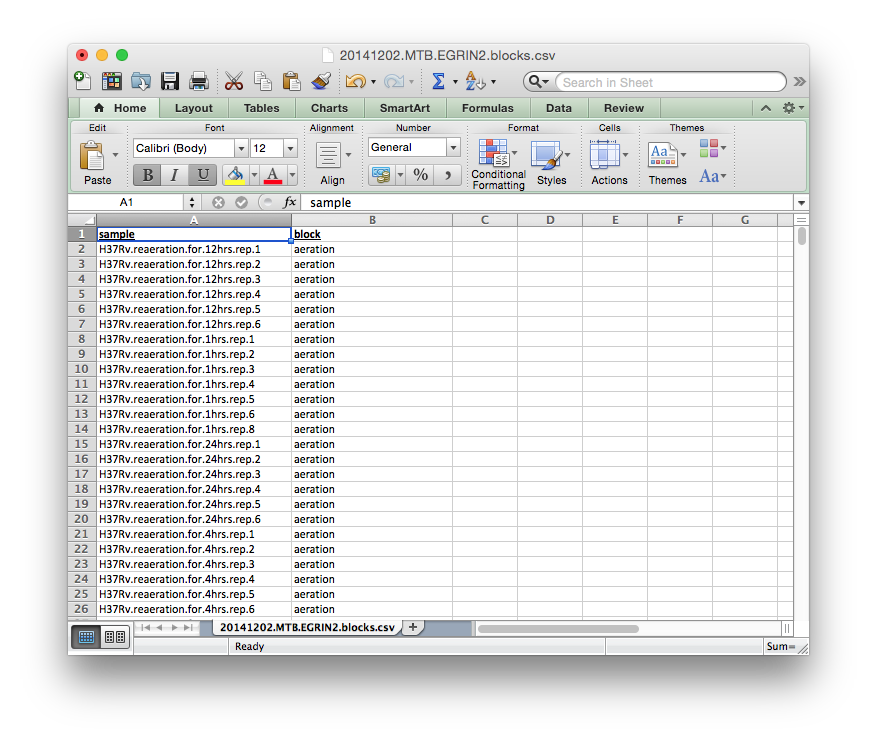
blocks.csv¶
A Tab-delimited or Comma-separated file containing the block membership for each experiment in the dataset as below.
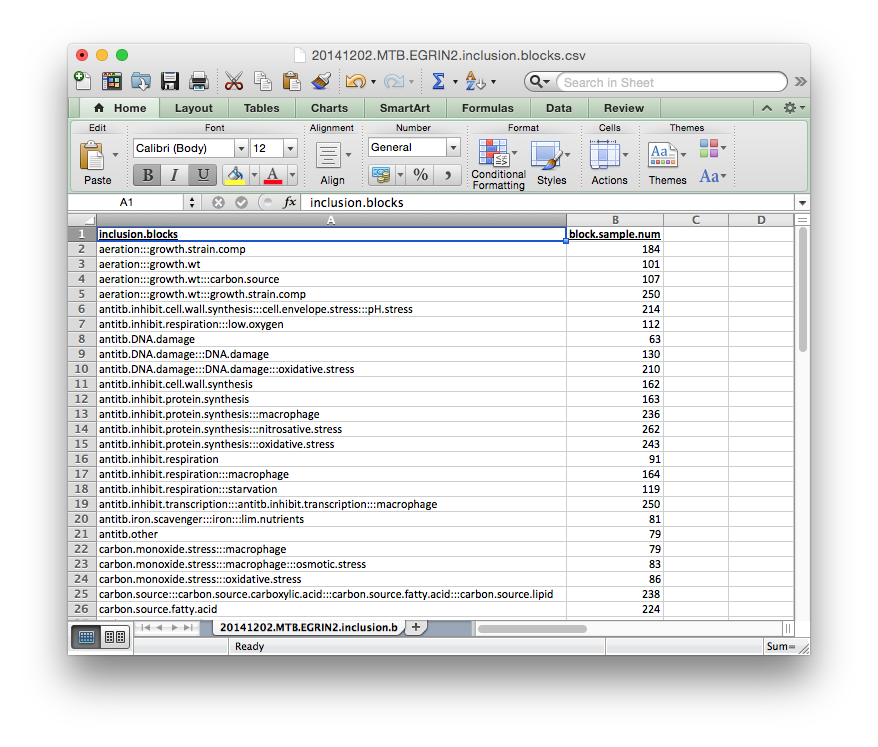
inclusion_blocks.csv¶
A Tab-delimited or Comma-separated file containing definitions for groups of blocks to be co-included in a cMonkey2 run, as defined above. These blocks are not strictly co-included in every cMonkey2 run, rather placement of a block into an inclusion block increases the likelihood that a particular block will be selected given that another block from its inclusion block has already been included in a particular cMonkey run.
Each group of blocks should be separated by :::, as indicated in the template below. The names of each block should be the same as those defined in blocks.csv.gz above.
The block.sample.num column is not required, but can be useful for evaluating the blocks.
exclusion_blocks.csv¶
A Tab-delimited or Comma-separated file containing definitions for groups of blocks to be co-excluded from a cMonkey2 run. Typically these blocks are defined for testing purposes. If they are not defined, several random exclusion blocks will be defined in order to evaluate model over-fitting (to be implemented)
The format for the exclusion blocks is the same as the inclusion blocks before. Each group of blocks should be separated by :::, as indicated in the template below. The names of each block refer to those defined in blocks.csv.gz above.
Again, the block.sample.num column is not required.
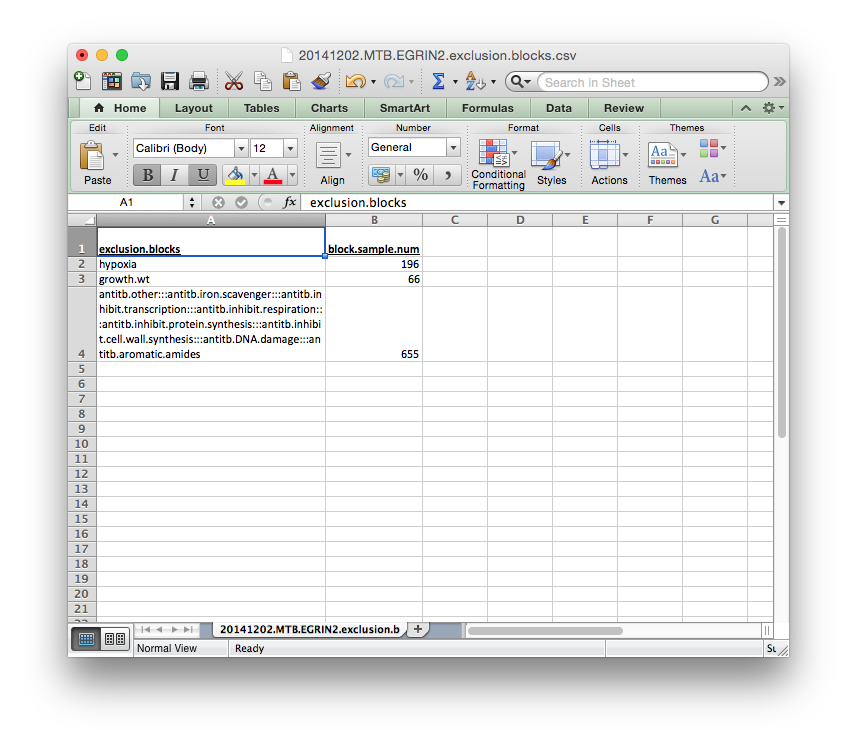
Each of these files need to be supplied by the user.
(Optional) Generate custom scoring pipeline and files¶
cMonkey2 can use a custom scoring pipeline. Currently, this is limited to set-enrichment.
In this example, we will generate an ensemble. We do so by providing several additional options to the egrin2-make_ensemble function, namely:
pipeline: a JSON file containing custom scoring pipeline. Currently only set-enrichment pipeline is supportedsetenrich: Name(s) of set enrichment ‘sets’ to include. Names should be comma separated.setenrich_files: Comma-separated files containing set-enrichment sets. Multiple files should be separated by a comma.
Users interested in building custom scoring pipelines or configuring set-enrichment should consult the cMonkey2 documentation.
STEP 2: Call egrin2-make_ensemble¶
Generating an ensemble configuration entails calling a single Python script. There are a number of required and optional parameters that can be passed to this command line function, which are described below:
$ egrin2-make_ensemble -h
usage: egrin2-make_ensemble [-h] --organism ORGANISM --ratios RATIOS
--targetdir TARGETDIR [--numruns NUMRUNS]
[--ncbi_code NCBI_CODE] [--mincols MINCOLS]
[--num_cores NUM_CORES] [--max_tasks MAX_TASKS]
[--user USER] [--csh] [--blocks BLOCKS]
[--inclusion INCLUSION] [--exclusion EXCLUSION]
[--pipeline PIPELINE] [--setenrich SETENRICH]
[--setenrich_files SETENRICH_FILES]
[--rsat_base_url RSAT_BASE_URL]
egrin2-make_ensemble - prepare cluster runs for Sun Grid Engine
optional arguments:
-h, --help show this help message and exit
--organism ORGANISM 3 letter organism code
--ratios RATIOS Path to ratios file
--targetdir TARGETDIR
Path to output directory
--numruns NUMRUNS Number of cMonkey2 runs to configure
--ncbi_code NCBI_CODE
NCBI organism code
--mincols MINCOLS Minimum number of experiments to include in a cMonkey2
run
--num_cores NUM_CORES
Number of cores on cluster to request
--max_tasks MAX_TASKS
Maximum number of jobs to be sent to the cluster at a
time
--user USER Cluster user name
--csh Flag to indicate C Shell
--blocks BLOCKS Path to block definitions
--inclusion INCLUSION
Path to inclusion block definitions
--exclusion EXCLUSION
Path to exclusion block definitions
--pipeline PIPELINE Path to scoring pipeline config file
--setenrich SETENRICH
Name(s) of set enrichment 'sets' to include. Names
should be comma separated.
--setenrich_files SETENRICH_FILES
Set enrichment files. File paths should be comma
separated.
--rsat_base_url RSAT_BASE_URL
Alternative RSAT base URL.
Here we will concentrate on the required arguments.
Required Arguments¶
organism: 3-letter organism coderatios: ratios file described abovetargetdir: location of a directory in which to configure the cMonkey2 runs
If you do not supply block files as described above, the experiments to include in each run will be choosen randomly. Several random exclusion_blocks will be defined for testing (currently not supported - block files must be supplied / ANB 03042015)
Here we will assume that the required files are in the local working directory. Furthermore, we will assume that the egrin2-make_ensemble is in the working directory and that all of the required modules are in your $PYTHONPATH.
For the following example, we will generate 5 cMonkey2 runs.
On the command line this would be called as follows:
$ egrin2-make_ensemble --organism mtu --ratios ratios.csv --targetdir mtu-ens-2014 --numruns 10 --blocks blocks.csv --inclusion inclusion_blocks.csv --exclusion exclusion_blocks.csv --pipeline setenrich_pipeline.json --setenrich chipseq,tfoe --setenrich_files ChIPSeq.csv,DE.csv --csh
If the script runs successfully, they should print the messages above, populate the mtu-ens-2014 directory with ratios-xxx.tsv files and config-xxx.ini files, as well as generate several report files in the parent directory. The ensemble report files contain information about the run composition, detailed for each report file below:
STEP 3: Evaluate ensembleReport files¶
ensembleReport_runs.csv
Global report of the ensemble run compositions.
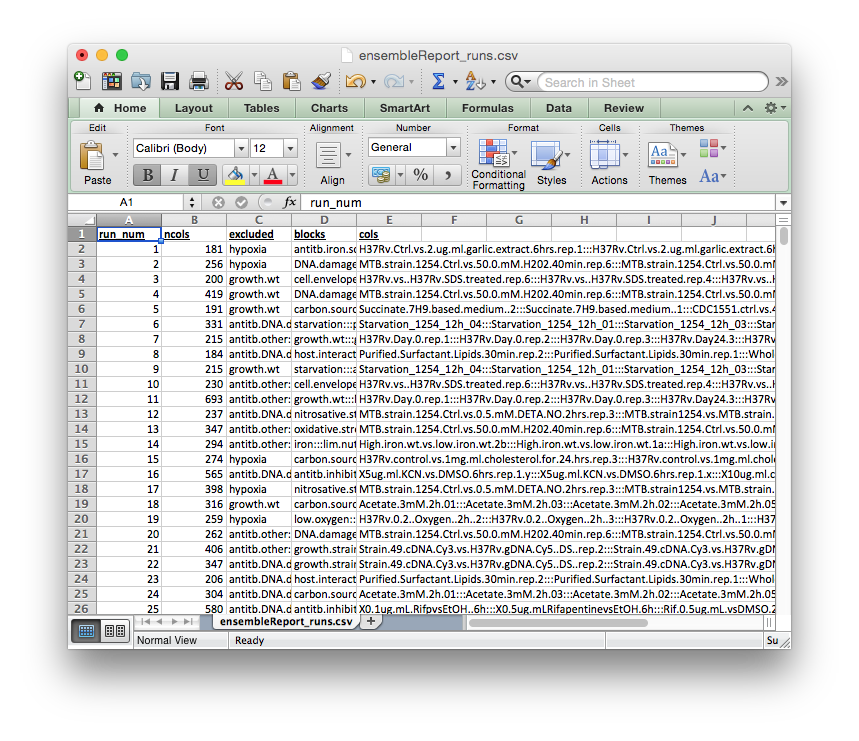
The columns of this report file signify:
run_num: cMonkey run numberncols: number of experiments included in the runexcluded:exclusion blockthat was excluded from the run (i.e., none of the conditions in this block will be in the run)blocks: blocks that were included in the runcols: names of experiments (from column names ofratios.csv) that were included in the run
ensembleReport_cols.csv¶
Extends blocks.csv. Reports how often each condition was included in the ensemble.
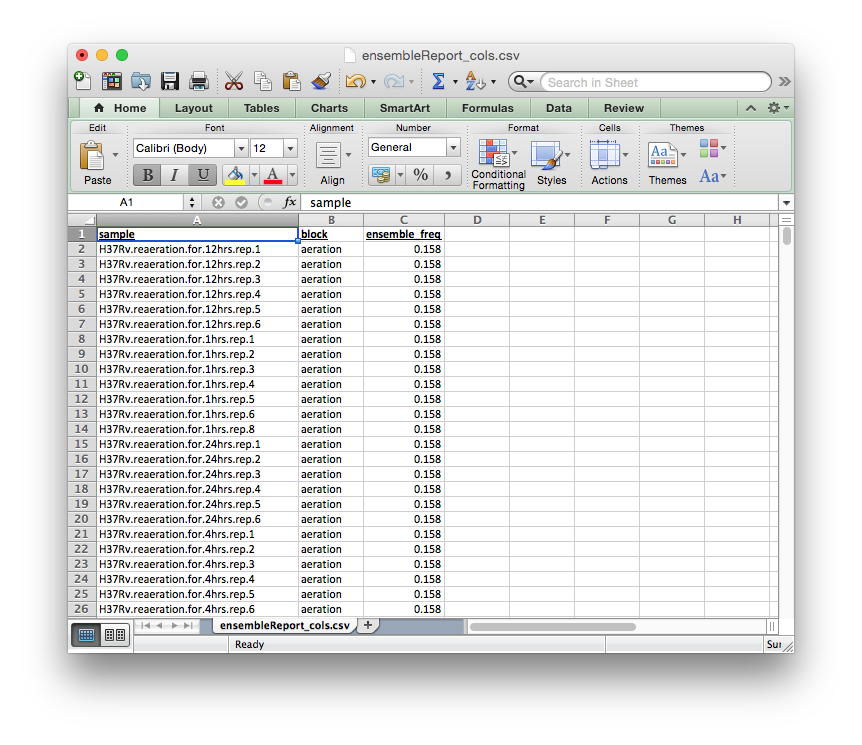
The columns of this report file signify:
sample: experiment name, fromblocks.csvblock:blockto which experiment belongs, fromblocks.csvensemble_freq: rate of inclusion in the ensemble
ensembleReport_blocks.csv¶
Reports how often each block was included in the ensemble.
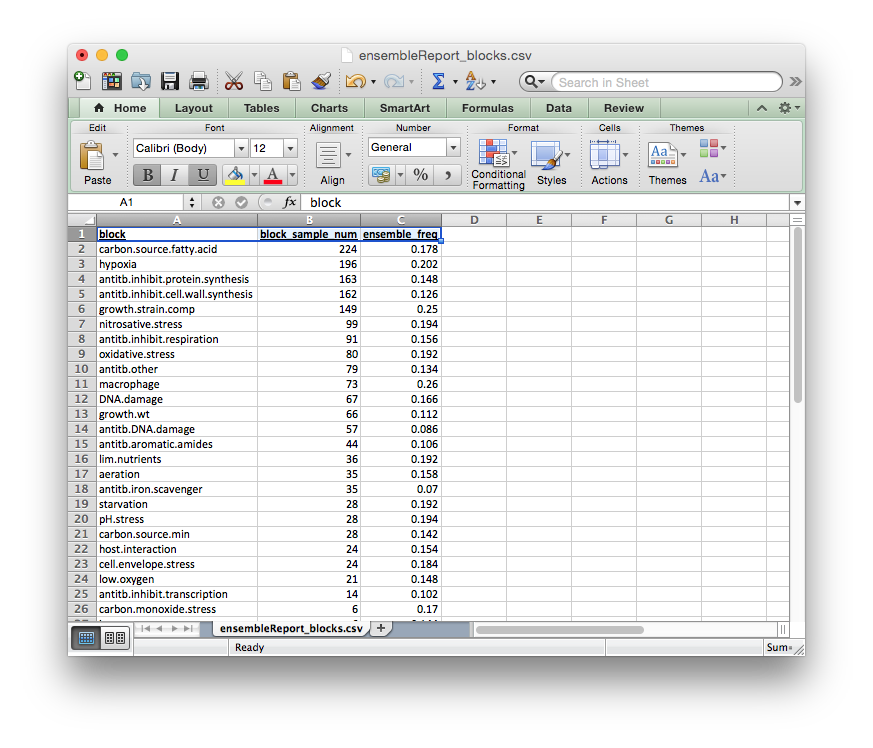
The columns of this report file signify:
block: block nameblock_sample_num: number of experiments annotated by this blockensemble_freq: rate of inclusion in the ensemble
ensembleReport_inclusionBlocks.csv¶
Reports how often each inclusion block was included in the ensemble.
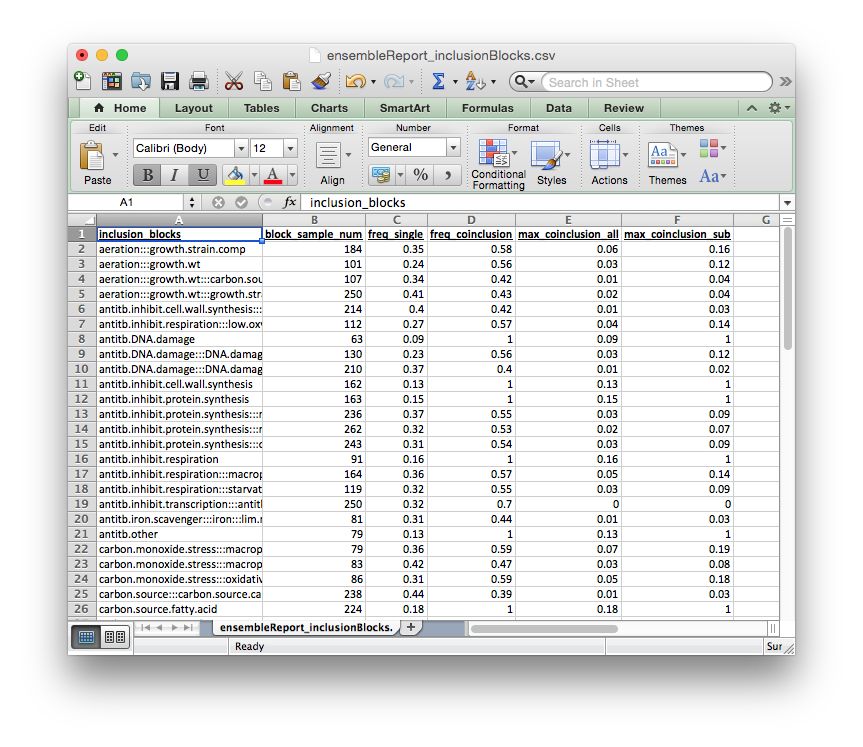
The columns of this report file signify:
inclusion_blocks: inclusion_block name. blocks separated by:::block_sample_num: number of experiments included in this inclusion blockfreq_single: rate at which a single block from this inclusion_block is included in the ensemblefreq_coinclusion: rate at which at least 2 of blocks from this inclusion_block are co-included in a single runmax_coinclusion_all: rate at which ALL of blocks from this inclusion_block are co-included in a single run across all ensemble runsmax_coinclusion_sub: rate at which ALL of blocks from this inclusion_block are co-included in a single run across all ensemble runs in which at least one of the blocks occurs
ensembleReport_exclusionBlocks.csv¶
Reports how often each exclusion block was excluded in the ensemble.
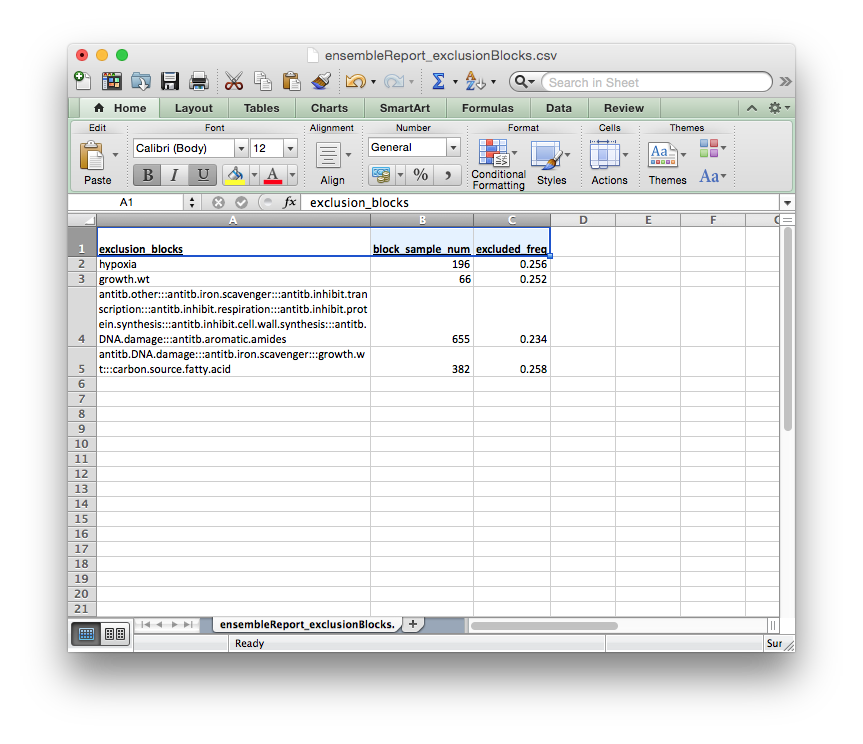
The columns of this report file signify:
exclusion_blocks: exclusion block nameblock_sample_num: number of experiments annotated by this exclusion blockexcluded_freq: rate of exclusion in the ensemble
STEP 4: Transfer to cluster and run cMonkey2¶
The entire targetdir (e.g. mtu-ens-2014 in our example) is now ready to be transfered to the cluster, where you can generate the ensemble by running <org>.sh, where <org> is the 3-letter organism code you provided. In our example case this would be mtu.sh.
Example for a SGE submission:
$ cd <target directory>
$ qsub mtu.sh